In 2014, Oregon Cryonics commissioned Advanced Neural Biosciences to obtain electron micrographs of brain tissue preserved using the VM-1 protocol developed by Dr. Yuri Pichugin while at the Cryonics Institute.
Methods
Five separate rat brains were prepared as follows:
0. Control, not cryoprotected or cooled. Only fixed.
1. VM-1 with sodium dodecylbenzenesulfonate (SDBS) a BBB modifier identified by Dr. Pichugin.
2. VM-1 with SDBS
3. VM-1
4. VM-1
Each of the five rats was euthanized via I.P. injection of 30% urethane (ethyl carbamate), then rapidly cooled by placing it in an ice bath in a refrigerator. At 20 deg C, the chest was opened and transcardial perfusion initiated. The open circuit consisted of a peristaltic pump, heat exchanger, and manometer. Arterial line pressure was stabilized at 100 mmHg.
100 ml of m-RPS-2 was perfused at 0 to 5 deg C.
The body was further cooled to 3 deg C.
The following cryoprotective agents (CPAs) were perfused at 0 to 5 deg C:
50 ml of 5% EG (in experiments 1 and 2, added 0.01% SDBS (0.005 g) to this step)
50 ml of 10% EG
50 ml of 20% EG
50 ml of 30% EG
VM-1 for the final step was chilled to -15 C in a beaker and, due to line warming and chiller settings, was perfused at -3 to -5 C.
200 ml of 70% VM-1
The brain was removed from the skull and transfered to a -130 C freezer for 24 hours. It was then allowed to thaw. The brain was sliced into cortical samples of approximately 3 mm. The CPA was unloaded from the tissue samples by placing sequentially in the following solutions:
30% EG (+ 300 mM mannitol)
20% EG (+ 300 mM mannitol)
10% EG (+ 300 mM mannitol)
5% EG (+ 300 mM mannitol)
m-RPS-2 (+ 300 mM mannitol)
m-RPS-2
The samples were placed in fixative solution (4% paraformaldehyde, 2% glutaraldehyde) for 16 hours, washed 3 x 30 mins in 0.1M phosphate buffer solution (PBS), and sent out for electron microscopy.
Results
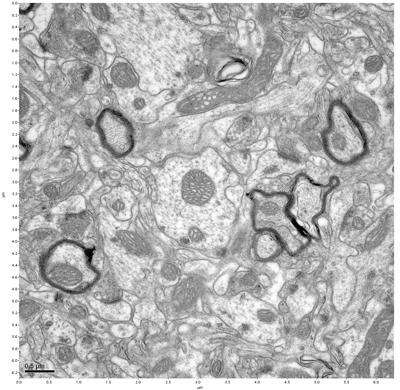
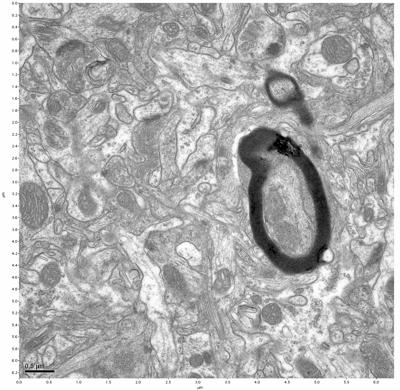
Figure 1. Control, image 1 of 2. Figure 2. Control, image 2 of 2.
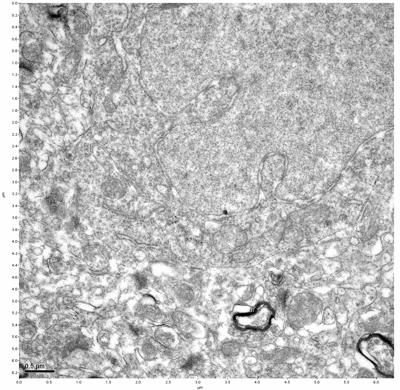
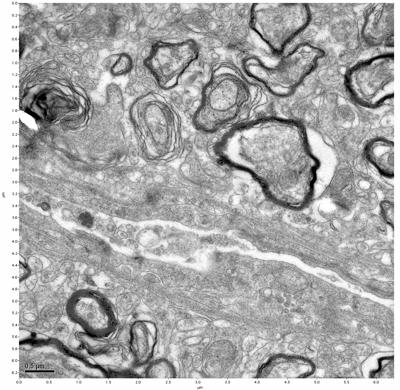
Figure 3. Sample one, image 1 of 4. Figure 4. Sample one, image 2 of 4.
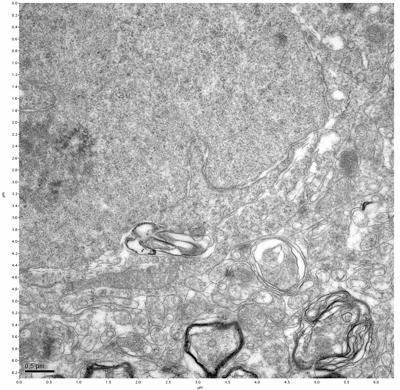
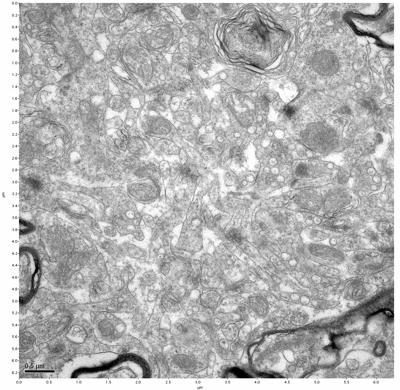
Figure 5. Sample one, image 3 of 4. Figure 6. Sample one, image 4 of 4.

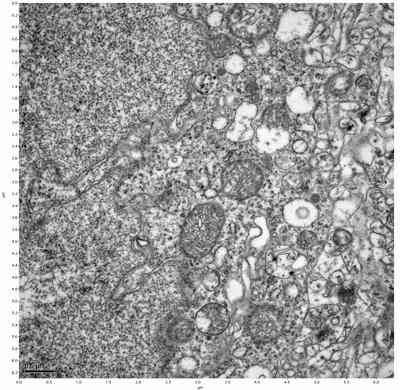
Figure 7. Sample two, image 1 of 6. Figure 8. Sample two, image 2 of 6.
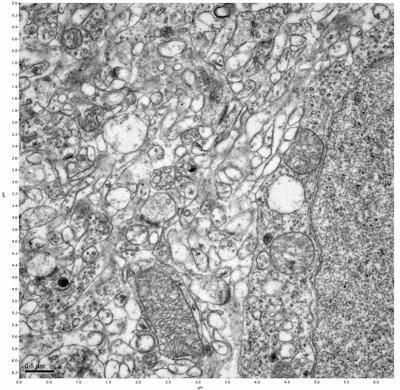
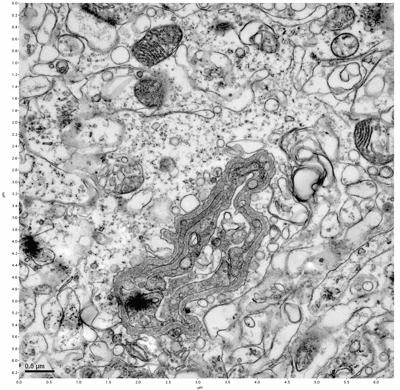
Figure 9. Sample two, image 3 of 6. Figure 10. Sample two, image 4 of 6.
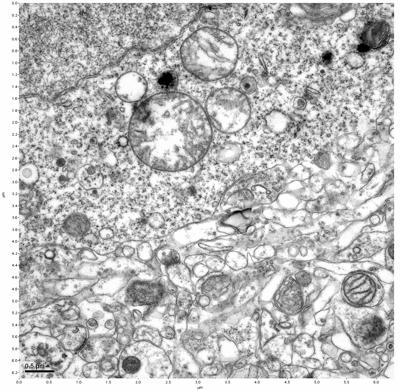
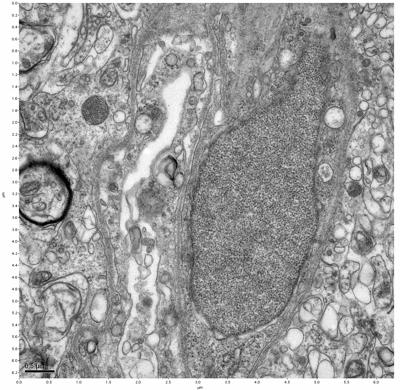
Figure 11. Sample two, image 5 of 6. Figure 12. Sample two, image 6 of 6.
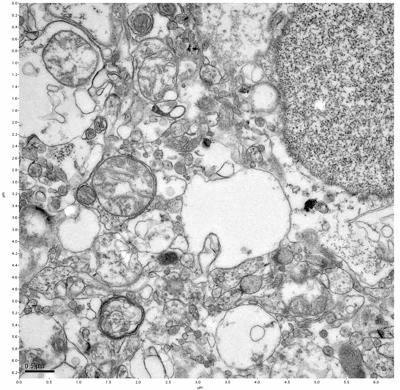
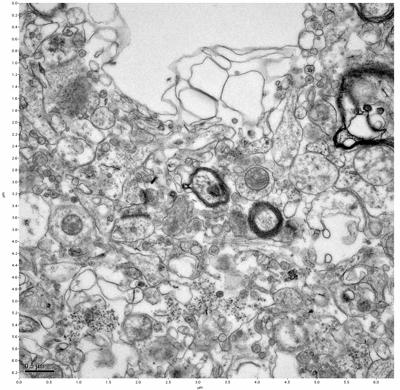
Figure 13. Sample three, image 1 of 4. Figure 14. Sample three, image 2 of 4.
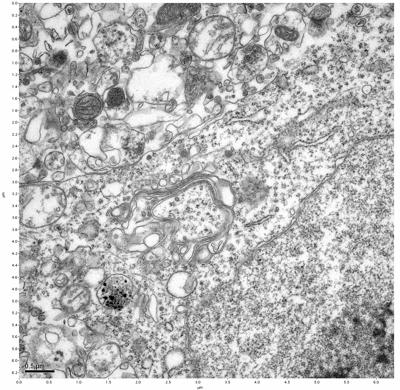
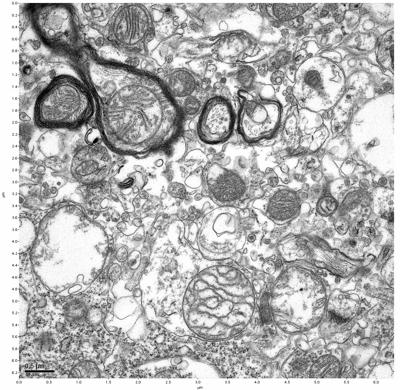
Figure 15. Sample three, image 3 of 4. Figure 16. Sample three, image 4 of 4.
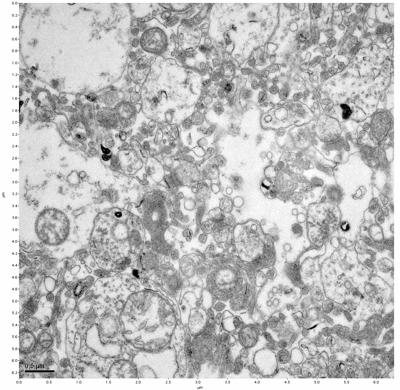
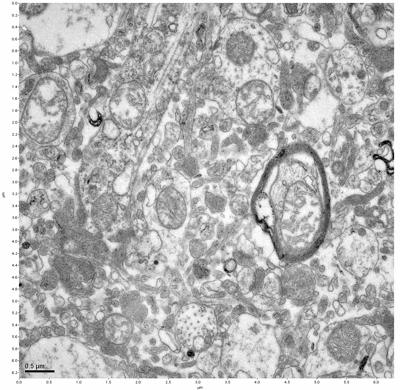
Figure 17. Sample four, image 1 of 4. Figure 18. Sample four, image 2 of 4.
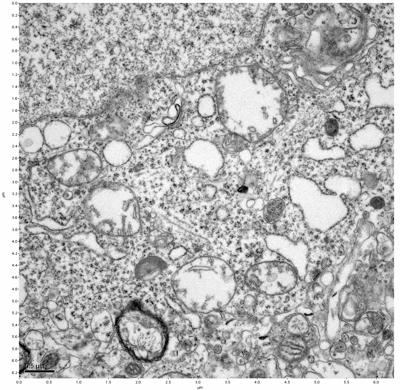
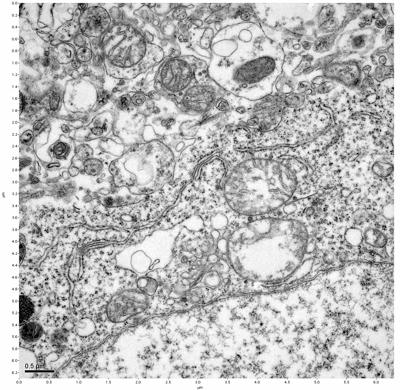
Figure 19. Sample four, image 3 of 4. Figure 20. Sample four, image 4 of 4.
This zip file
ElectronMicrographs.zip (87 MB)
contains the EMs above at slightly higher resolution as well as some additional EMs.
The cryopreserved samples display some loss of structure. The control sample clearly has better overall ultrastructural preservation relative to the four crypopreserved samples. Samples 3 & 4 are in slightly worse condition than samples 1 & 2, showing open debris fields of a few microns in diameter (for example, sample 4 image 1), but are otherwise of comparable quality. Mitochondria in all four samples notably appear degraded, with fewer or no cristae. Cellular membranes and endomembrane structures are frequently degraded and sometimes missing. Synapses and synaptic vesicles are observed more frequently in the control than in any of the samples. Although many original structures are identifiable, the cytoplasm itself shows a loss of complexity and organization, with a more granular and open appearance. The cryopreserved samples do not display any evidence of freezing or large-scale distortion.
The purpose of the SDBS was to impede severe dehydration shrinkage. As expected, the SDBS brains did not shrink at all. The VM-1 brains (3 & 4) experienced severe shrinkage of nearly 50%. This shrinkage is not evident in the images because the CPA was unloaded, and the rehydration returned the samples to a state closer to their original morphology.
Discussion
These electron micrographs demonstrate clear preservation of significant structure as well as a certain amount of damage. Some of the damage can be attributed to thawing, unloading the CPA, and processing for electron microscopy. Cryopreserved tissue which had not been thawed and processed would probably be in better condition than the tissue in these images.
This was an ideal lab setting which has only a limited resemblance to a human case. Most importantly, these were healthy brains, whereas human cases would nearly always present with severely altered physiology. The brains were much smaller than human brains. Perfusion times were correspondingly shorter, subjecting the tissue to less damage. Cooldown times were able to be much quicker, both for the initial hypothermia as well as during subzero cooling. No time was added to simulate legal delays in beginning procedures immediately post pronouncement. Human cases are rarely as ideal as this.
SDBS was effective in preventing shrinkage in brains with no ischemia. This resulted in slightly better results, but since most human patients suffer ischemia, the relevance is not clear. We are not adding SDBS to our protocol at this time because the effect is too strong. We would prefer some mild shrinkage because it provides stability and facilitates vitrification.
The clear preservation of structure in these cryopreserved samples provides very strong evidence for the assertion that at least part of the connectome is intact and recoverable in the future.
